Structures
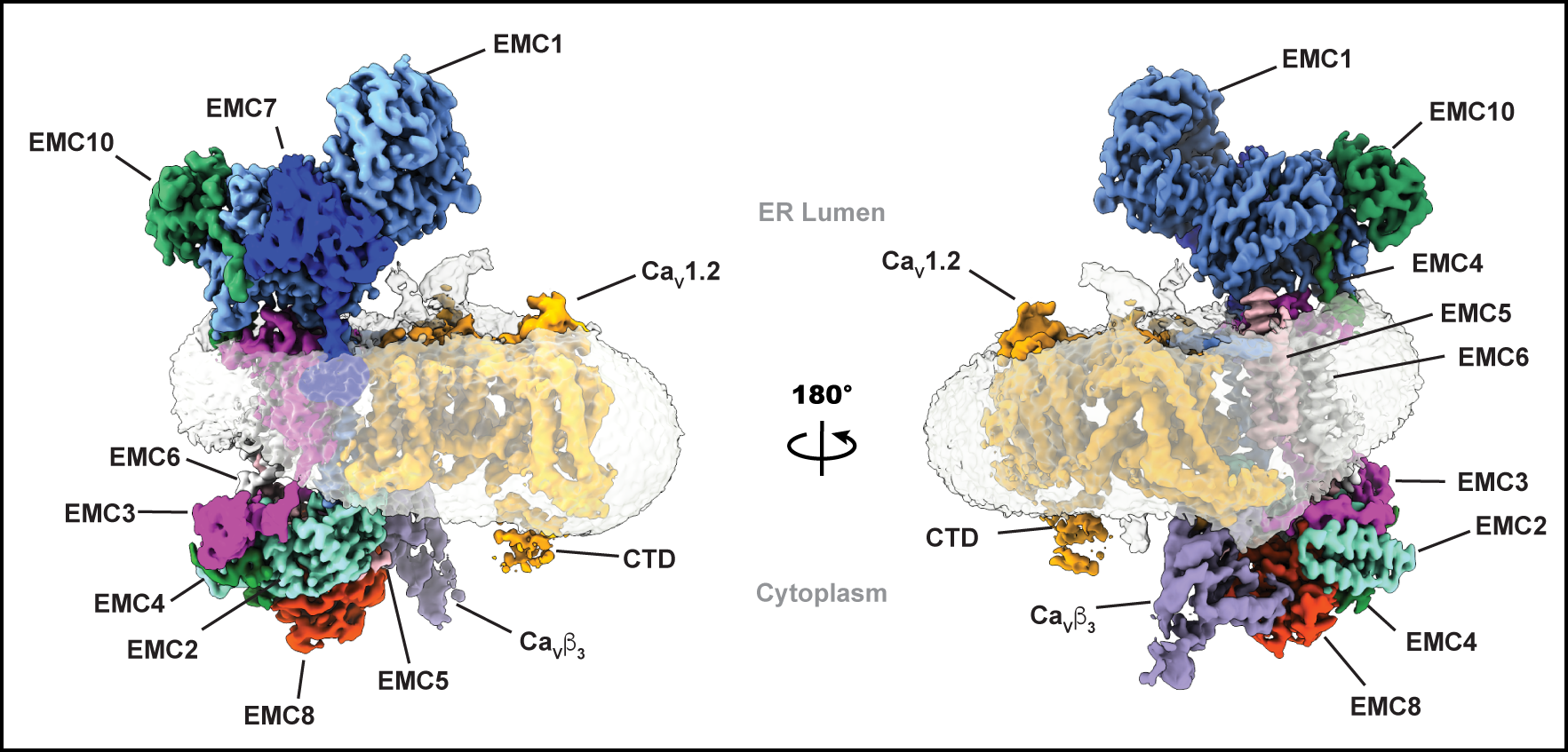
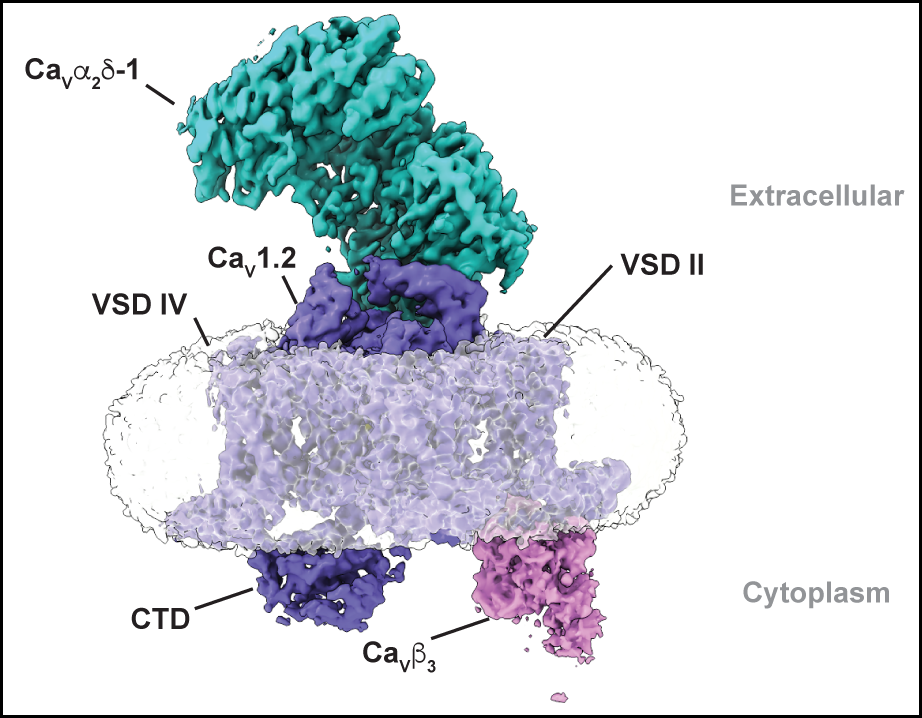
EMC:Cavcomplex
Cav1.2 structures
Cav1.2(ΔC):Cavβ3Cavα2δ-1 complexed with L-Leu
PDB: 8EOG
EMDB:EMD-28375, EMD-28561, EMD-28564, EMD-40561
Cav1.2(ΔC):Cavβ3Cavα2δ-1 complexed with gabapentin
PDB: 8FD7
EMDB:EMD-29004, EMD-29007, EMD-29015
EMC chaperone-CaV structure reveals an ion channel assembly intermediate. Chen Z, Mondal, A., Aberemane-Ali, F., Jang, S., Niranjan, S., Montaño, J.L., Zaro, B.W., Minor, D.L., Jr. Nature 619 410-419 (2023)
Structural basis for CaVɑ 2δ:gabapentin binding. Chen Z, Mondal A, and Minor, D.L., Jr. Nature Struct. & Mol. Biol. 30 735-739 (2023)
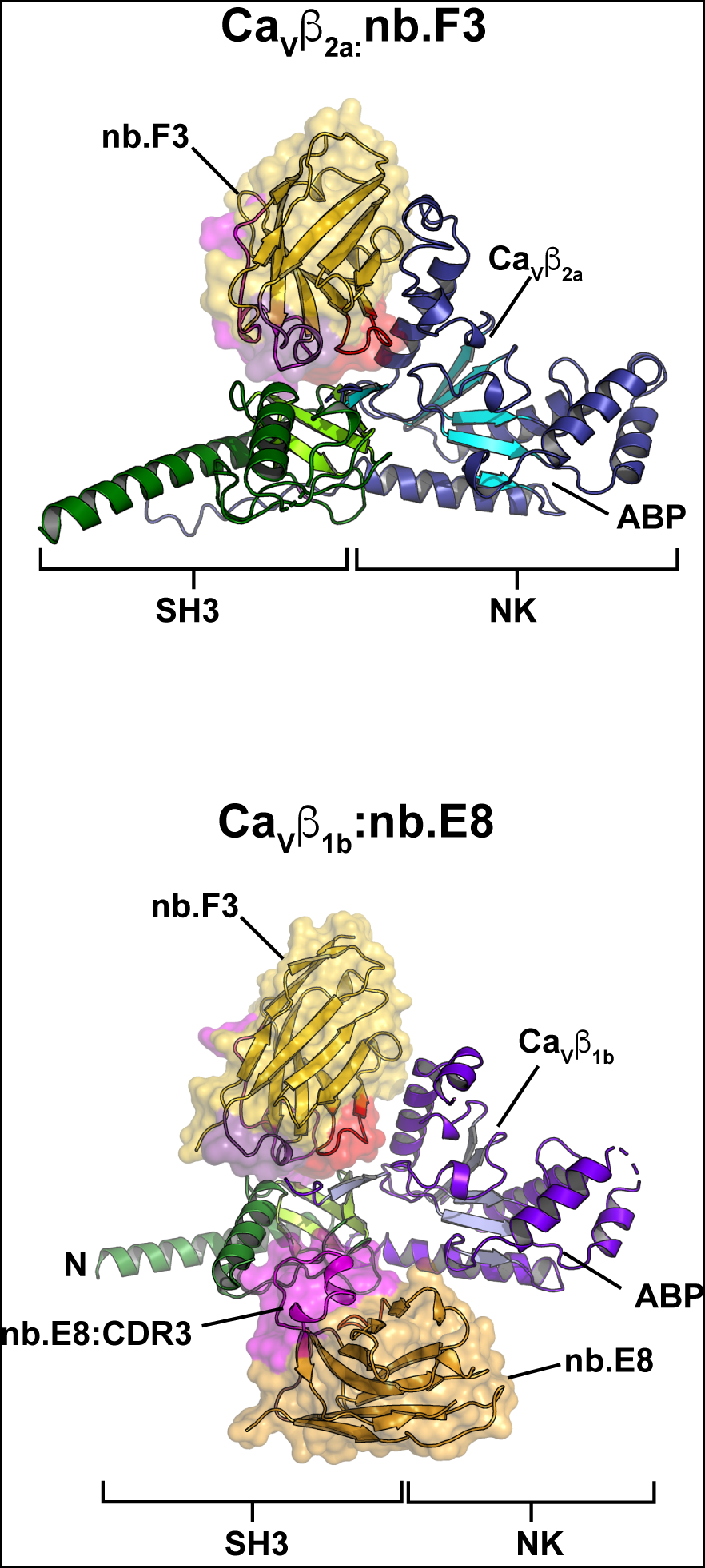
Nanobody: Cavβ complexes
Cavβ1b:nb.E8
PDB: 8DAM
Cavβ2a:nb.F3
PDB: 8EOE
Selective posttranslational inhibition of CaVβ1-associated voltage-dependent calcium channels with a functionalized nanobody. Morgenstern, T., Nirwan, N., Hernandez-Ochoa, E., Bibollet, H., Choudhury, P., Laloudakis, J., Ben-Johny, M., Bannister, R., Schneider, M., Minor, D.L., Jr., and Colecraft, H.M. Nature Communications 13:7556 (2022)
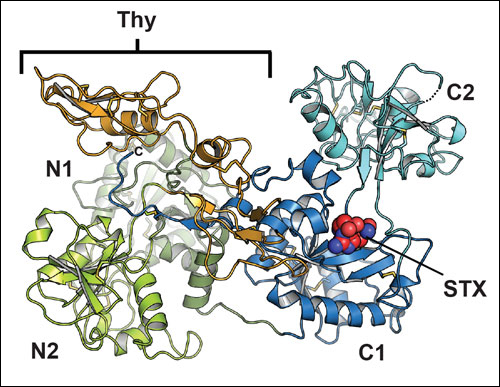
Saxiphilin variants and complexes
RcSxph-Y558A
PDB: 8D6P
RcSxph-Y558A:STX
PDB: 8D6T
RcSxph-Y558I
PDB: 8D6Q
RcSxph-Y558I:STX
PDB: 8D6T
RcSxph:F-STX (Flourescein-STX)
PDB: 8D6P
High Himalaya Frog (N. parkeri), saxiphilin (NpSxph)
PDB: 8D6G
NpSxph:STX
PDB: 8D6M
NpSxph:F-STX
PDB: 8D6O
Definition of a saxitoxin (STX) binding code enables discovery and characterization of the Anuran saxiphilin family. Chen, Z., Zakrzewska, S., Hajare, H., Alvarez-Bullya, A., Abderemane-Ali, F., Bogan, M., Ramirez, D., O’Connell, L.A., Du Bois, J., and Minor, D.L., Jr. Proceedings of the National Academy of Sciences, USA 119:e2210114119 (2022)

Non-canonical BacNaV pore domain structures
NavAb1p (DM-detergent) ‘Inside-out’
PDB: 7PGG
NavAb1p (bicelles) ‘Inside-out’
PDB: 7PGI
CavSp1p (bicelles) ‘PeripheralSF’
PDB: 7PGF
NavAe1/Sp1CTDp (DDM-detergent) ‘Inside-out’
PDB: 7PGH
Fab complexes
NavAe1/Sp1CTDp:SAT09 complex
PDB: 7PGB
NavAe1/Sp1CTDp:ANT05 complex
PDB: 7PG8
Quaternary structure independent folding of voltage-gated ion channel pore domain subunits. Arrigoni, C., Lolicato, M., Shaya, D., Rohaim, A., Findeisen, F., Fong, L.-K., Colleran, C.M., Dominik, P., Kim, S.S., Schuermann, J., DeGrado, W.F., Grabe, M., Kossiakoff, A.A., and Minor, D.L., Jr. Nature Structural and Molecular Biology 29: 537-548 (2022)

K2P2.1 (TREK-1) in varied potassium concentrations
K2P2.1 (TREK-1), 0 mM [K+]
PDB: 6W7B
K2P2.1 (TREK-1), 1 mM [K+]
PDB: 6W7C
K2P2.1(TREK-1), 10 mM [K+]
PDB: 6W7D
K2P2.1 (TREK-1), 30 mM [K+]
PDB: 6W7E
K2P2.1 (TREK-1), 50 mM [K+]
PDB: 6W82
K2P2.1 (TREK-1), 100 mM [K+]
PDB: 6W83
K2P2.1 (TREK-1), 200 mM [K+]
PDB: 6W84
K2P2.1 (TREK-1):ML335 complex, 0 mM [K+]
PDB: 6W8F
K2P2.1 (TREK-1):ML335 complex, 1 mM [K+]
PDB:6W8C
K2P2.1 (TREK-1):ML335 complex, 10 mM [K+]
PDB: 6W8A
K2P2.1(TREK-1):ML335 complex, 30 mM [K+]
PDB: 6W88
K2P2.1 (TREK-1):ML335 complex, 50 mM [K+]
PDB: 6W87
K2P2.1 (TREK-1):ML335 complex, 100 mM [K+]
PDB: 6W86
K2P2.1 (TREK-1):ML335 complex, 200 mM [K+]
PDB: 6W85
K2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions. Lolicato, M., Natale, A.M., Abderemane-Ali, F., Crottes, D., Capponi, S., Duman, R. Wagner, A., Rosenberg, J.M., Grabe, M., Minor, D.L. Jr. Science Advances 6 eabc9174 (2020)

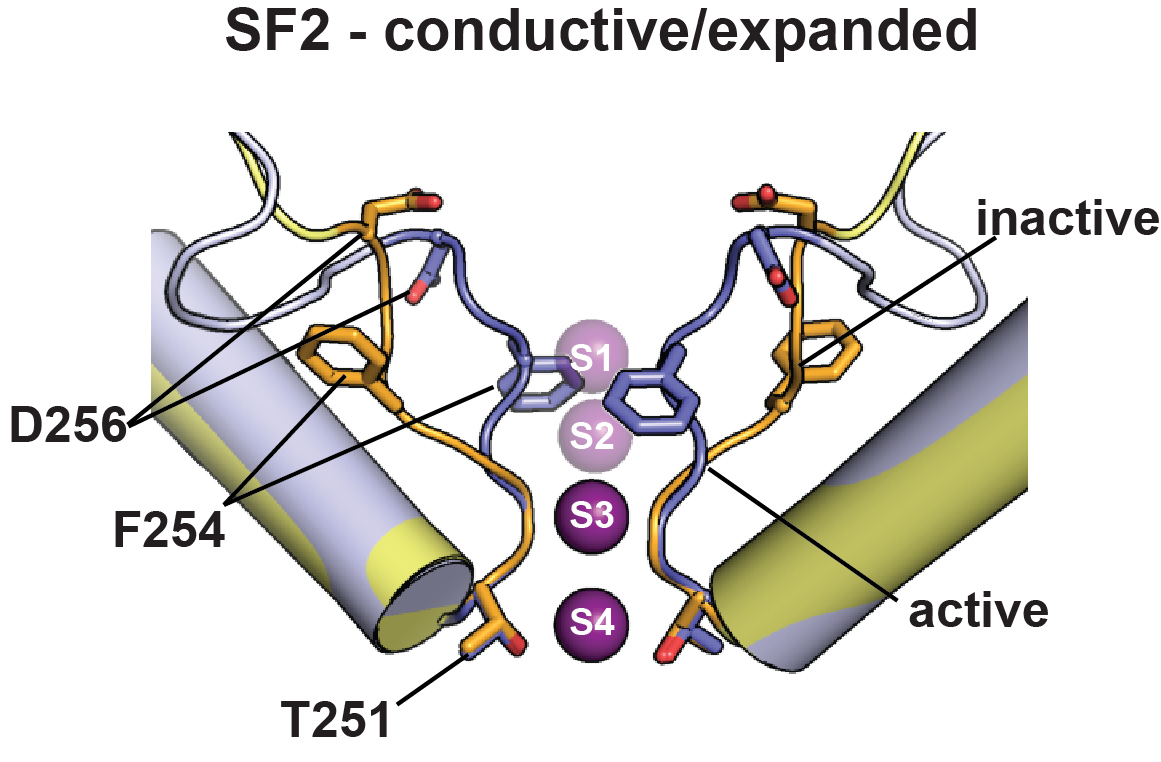
K2P2.1 (TREK-1):Polynuclear ruthenium amine complexes
K2P2.1 (TREK-1) I110D
PDB: 6V36
K2P2.1 (TREK-1) I110D:Ruthenium red
PDB: 6V3I
K2P2.1 (TREK-1) I110D:Ruthenium red:ML335
PDB: 6V37
K2P2.1 (TREK-1) I110D:Ru360
PDB: 6V3C
Polynuclear Ruthenium Amines Inhibit K2P Channels via a "Finger in the Dam" Mechanism. Pope, L., Lolicato, M., Minor, D.L. Jr. Cell Chemical Biology 27: 511-524 (2020)

Saxiphilin and Saxiphilin:STX complexes
Saxiphilin
Apo-Saxiphilin
PDB ID: 6O0D
Saxiphilin: STX complexes
Saxiphilin:STX complex (soaked)
PDB ID: 6O0E
Saxiphilin:STX complex (co-crystal)
PDB ID: 6O0F
Structure of the saxiphilin:saxitoxin (STX)complex reveals a convergent molecular recognition strategy for paralytic toxins. Yen, T.-J., Lolicato, M., Thomas-Tran, R., Du Bois, J., and Minor, D.L. Jr., Science Advances 5 eaax2650 (2019) PMID: 31223657 PMCID: PMC6584486 View Press
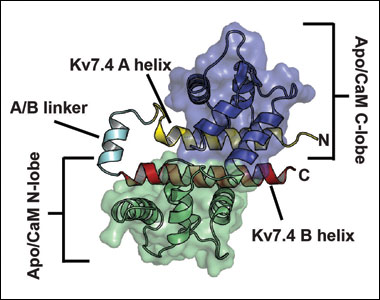
CaM:Kv7.4 AB domain complexes & CaM:Kv7.5 AB domain complex
CaM:Kv7.4 AB domain complexes:
Apo/CaM:Kv7.4 AB domain complex
PDB ID: 6B8L
Ca2+/CaM:Kv7.4 AB domain complex, 1 mM CaCl2 soak
PDB ID: 6B8M
Ca2+/CaM:Kv7.4 AB domain complex, 10 µM CaCl2 soak
PDB ID: 6B8N
Mg2+/CaM:Kv7.4 AB domain complex
PDB ID: 6B8P
CaM:7.5 AB domain complex:
Mg2+/CaM:Kv7.5 AB domain complex
PDB ID: 6B8Q
A Calmodulin C-Lobe Ca2+-Dependent Switch Governs Kv7 Channel Function. Chang, A., Abderemane-Ali, F., Hura, G.L., Rossen, N.D., Gate, R.E., Minor, D.L., Jr., Neuron 97 836-852 (2018) View Video Abstract PMID:29429937 PMCID: PMC5823783
TMEM16A calcium-activated chloride channel
Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc
PDB ID: 6BGI
Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG
PDB: 6BGJ
Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Dang S., Feng S., Tien J., Peters C.J., Bulkley D., Lolicato M., Zhao J., Zuberbühler K., Ye W., Qi L., Chen T., Craik C.S., Nung Jan Y., Minor D.L. Jr, Cheng Y., Yeh Jan L., Nature 552 426-429 (2017) PMID: 29236684 PMCID: PMC5750132
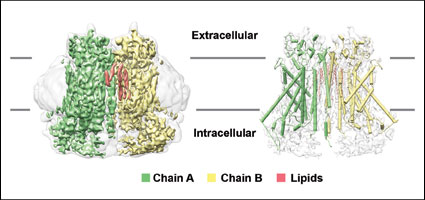
Coagulation Factor XI:antibody complex
FXIa/ DEF Fab complex coordinates
PDB: 6AOD
Structural basis for activity and specificity of an anticoagulant anti-FXIa monoclonal antibody and a reversal agent. Ely, L., Lolicato, M. David, T., Lowe, K., Kim, Y.C., Samuel, D., Bessette, P., Garcia, J.L., Mikita, T., Minor, D.L. Jr., Coughlin, S.R., Structure 26187-198 (2018)

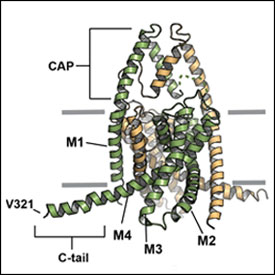
Mouse K2P2.1 (TREK-1)
K2P2.1 (TREK-1)
PDB ID: 5VK5
Mouse K2P2.1 (TREK-1): Activator Complexes
ML335 complex
PDB ID: 5VKN
ML402 complex
PDB ID: 5VKP
K2P2.1(TREK-1):activator complexes reveal a cryptic selectivity filter binding site. Lolicato, M., Arrigoni, C., Mori, T., Sekioka, Y., Bryant, C., Clark, K.A., Minor, D.L., Jr., Nature 547 364-368 (2017) PMID: 28693035 PMCID: PMC5778891
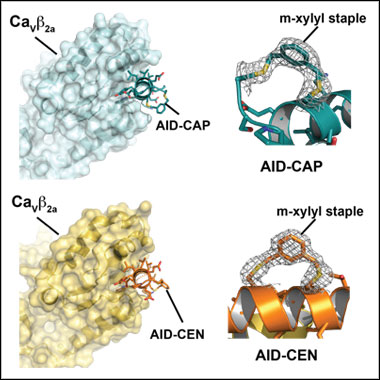
CaVβ: Stapled peptide complexes
AID-CAP PDB ID: 5V2P
AID-CEN PDB ID: 5V2Q
Stapled voltage-gated calcium channel (CaV) α-Interaction Domain (AID) peptides act as selective protein-protein interaction inhibitors of CaV Function. Findeisen, F., Campiglio, M., Jo, H., Abderemane-Ali, F., Rumpf, C.H., Pope, L., Rossen, N.D., Flucher, B.E., DeGrado, W.F., and Minor D.L., Jr., ACS Chemical Neuroscience 8 1313-1326 (2017) PMID: 28278376 PMCID: PMC5481814
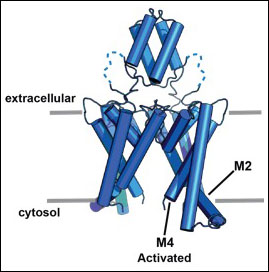
Human K2P4.1 (TRAAK)
G124I
PDB ID: 4RUE
W262S
PDB ID: 4RUF
Transmembrane helix straightening and buckling underlies activation of mechanosensitive and thermosensitive K2P channels.
Lolicato, M., Riegelhaupt, P.M., Arrigoni, C., Clark, K.A., Minor, D.L., Jr., Neuron 84 1198-1212 (2014) PMID: 25500157 PMCID: PMC4270892
NaVAe1p (Alkalilimnicola ehrlichii)
PDB IDs: 4LTO, 4LTP, 4LTQ, and 4LTR
Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
Shaya, D., Findeisen, F., Abderemane-Ali, F., Arrigoni, C., Wong, S., Reddy Nurva, S., Loussouarn, G., and Minor, D.L., Jr., Journal of Molecular Biology 426 476-483 (2014) PMID: 24120938 PMCID: PMC3947372
» See Cover
PDB IDs: NaVAe1p 2.95Å: 5HK7, 5IWO, 5IWN, 3G neck mutant: 5HJ8, 5HK6, 7G neck mutant: 5HKD
Unfolding of a temperature-sensitive domain controls voltage-gated channel activation.
Arrigoni, C., Rohaim, A., Shaya, D., Findeisen, F., Stein, R.A. Nurva, S.R., Mishra, S., Mchaourab, H.S., and Minor, D.L., Jr., Cell 164 922‑936 (2016)

Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase
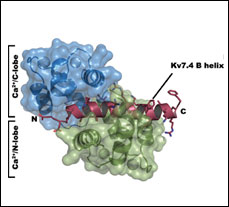
Ca2+CaM:Kv7.4 (KCNQ4) B-helix complex
PDB ID: 4GOW
Structure of a Ca2+/CaM:Kv7.4 (KCNQ4) B-helix complex provides insight into M-current modulation.
Xu, Q., Chang, A., Tolia, A., and Minor, D.L., Jr., Journal of Molecular Biology 425 378-394 (2013) PMID: 23178170 PMCID:PMC3540129
Ca2+/CaM-CaV1.2 PreIQ-IQ domain complex
PDB ID: 3OXQ
Multiple C-terminal tail Ca2+/CaMs regulate CaV1.2 function but do not mediate channel dimerization.
Kim, E., Rumpf, C., Van Petegem, F., Arant, R.J., Findeisen, F., Cooley, E., Isacoff, E., Minor, Jr., D.L., The EMBO Journal 29, 3924-3938 (2010)

CaBP1 and CaBP1 K130A
CaBP1
PDB ID: 3OX5
CaBP1 K130A
PDB ID: 3OX6
Structural Basis for the Differential Effects of CaBP1 and Calmodulin on CaV1.2 Calcium-Dependent Inactivation.
Findeisen, F., Minor, Jr., D.L., Structure 18, 1617–1631 (2010)

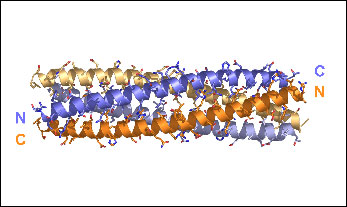
A trimeric form of the KV7.1 A domain Tail
Xu, Q., Minor, Jr., D.L., Crystal structure of a trimeric form of the KV7.1 (KCNQ1) A-domain tail coiled-coil reveals structural plasticity and context dependent changes in a putative coiled-coil trimerization motif. Protein Science 18:2100—2114 (2009)
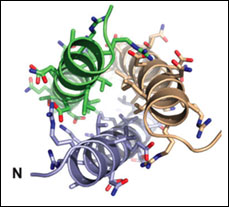
TRPM7 Assembly Domain
PDB ID: 3E7K
Fujiwara, Y., Minor, Jr., D.L. X-ray Crystal Structure of a TRPM Assembly Domain Reveals an Antiparallel Four-stranded Coiled-coil.
J. Mol. Biol. 383, 854-870 (2008).
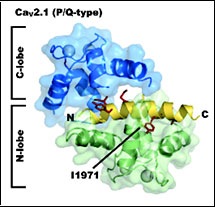
CaV 2.1 IQ domain-Ca2+/Calmodulin Complex
PDB ID: 3DVM
Kim, E.Y., Rumpf, C.H., Fujiwara, Y., Cooley, E.S., Van Petegem, F., and Minor, Jr., D.L. Structures of CaV2 Ca2+/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure 16, 1455-1467 (2008).
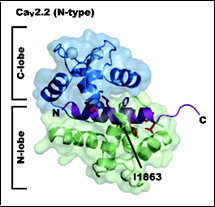
CaV 2.2 IQ domain-Ca2+/Calmodulin Complex
Kim, E.Y., Rumpf, C.H., Fujiwara, Y., Cooley, E.S., Van Petegem, F., and Minor, Jr., D.L. Structures of CaV2 Ca2+/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation. Structure 16, 1455-1467 (2008).
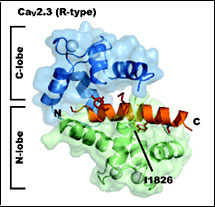
CaV 2.3 IQ domain-Ca2+/Calmodulin Complex
PDB ID: 3DVK
Kim, E.Y., Rumpf, C.H., Fujiwara, Y., Cooley, E.S., Van Petegem, F., and Minor, Jr., D.L. Structures of CaV2 Ca2+/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation. Structure 16, 1455-1467 (2008).
Kv7.4 (KCNQ4) assembly specificity domain
2OVC Kv7.4 (KCNQ4) Assembly Specificity Domain
PDB ID: 2OVC
Structural insight into KCNQ (Kv7) channel assembly and channelopathy.
Howard, R.J., Clark, K.A., Holton, J.M., and Minor, Jr., D.L., Neuron 53 663-675 (2007)
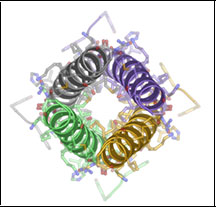
KChIP1/Kv4.3 T1 domain complex
2I2R KChIP1/Kv4.3 T1 Domain Complex
PDB ID: 212R
Three-dimensional structure of the KChIP/Kv4.3 T1 domain complex reveals a cross-shaped octamer. Pioletti, M., Findeisen, F., Hura, G.L., and Minor, Jr., D.L., Nature Structural & Molecular Biology 13 987 995 (2006)
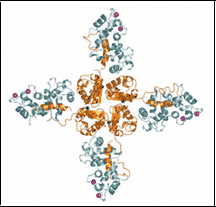
Voltage-gated Potassium Channel T1 assembly domain
1QDW Kv1.2 T1 domain 33-119
PDB ID: 1QDW
1QDV Kv1.2 T1 domain 33-131
PDB ID: 1QDV
1DSX KV1.2 T1 domain33-131 T46V mutant
PDB ID: 1DSX
The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel. Minor, Jr., D.L., Lin, Y.F, Mobley, B.C., Avelar, A., Jan, Y.N., Jan, L.Y. and Berger, J.M., Cell 102 657-670 (2000)
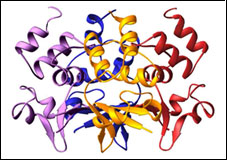
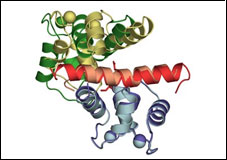
Voltage-gated Calcium Channel β-subunit
1T0H CaVβ2a
PDB ID: 1TOH
1T0J CaVβ2a - CaV1.2 AID complex
PDB ID: 1TOJ
Structure of a complex between a voltage gated calcium channel beta-subunit and an alpha-subunit domain. Van Petegem, F., Clark, K.A., Chatelain, F.C., and Minor, Jr., D.L., Nature 429 671-675 (2004)
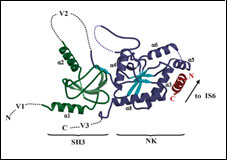
CaV1.2 IQ domain-Ca2+/Calmodulin Complex
2BE6 CaV1.2 IQ domain-Ca2+/CaM complex
PDB ID: 2BE6
Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Van Petegem, F, Chatelain, F.C., Minor, Jr., D.L., Nature Structural & Molecular Biology 12 1108-1115 (2005)