
Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Torosyan H, Paul MD, Forget A, Lo M, Diwanji D, Pawłowski K, Krogan NJ, Jura N, Verba KA.
Nature Communications 2023 Jun 19;14(1):3543
Deposited structure of the PEAK3 pseudokinase dimer
PDB: 8DS6Deposited structure of the PEAK3/14-3-3 complex
PDB: 8DP5
Trapping Tribbles: Nanobody-assisted structure of the TRIB2 pseudokinase.
Santana FR, Linossi EM, Jura N
Structure 2022 Nov 3;30(11):1465-1467
Preface to Special Issue of Methods in Enzymology on Pseudokinases.
Jura N, Murphy JM.
Methods in Enzymology 2022;667:xxi-xxiv
Link to the Special Issue of Methods in Enzymology: Pseudokinases
Efficient expression, purification, and visualization by cryo-EM of unliganded near full-length HER3.
Diwanji D, Trenker R, Jura N, Verba KA.
Methods in Enzymology 2022;667:611-632
An effective strategy for ligand-mediated pulldown of the HER2/HER3/NRG1β heterocomplex and cryo-EM structure determination at low sample concentrations.
Trenker R, Diwanji D, Verba KA, Jura N.
Methods in Enzymology 2022;667:633-662
CNPY4 inhibits the Hedgehog pathway by modulating membrane sterol lipids.
Lo M, Sharir A, Paul M, Torosyan H, Agnew C, Li A, Neben C, Marangoni P, Xu L, Raleigh DR, Jura N, Klein OD.
Nature Communications 2022 May 3; 13(1): 2407
Piquing our interest: Insights into the role of PEAK3 in signaling and disease.
Paul MD, Torosyan H, Jura N.
Science Signaling 2022 Feb 22;15(722):eabm9396
Extensive conformational and physical plasticity protects HER2-HER3 tumorigenic signaling.
Campbell MR, Ruiz-Saenz A, Zhang Y, Peterson E, Steri V, Oeffinger J, Sampang M, Jura N, Moasser MM.
Cell Reports 2022 Sep 6;40(10):111338
Targetable HER3 functions driving tumorigenic signaling in HER2-amplified cancers.
Campbell MR, Ruiz-Saenz A, Peterson E, Agnew C, Ayaz P, Garfinkle S, Littlefield P, Steri V, Oeffinger J, Sampang M, Shan Y, Shaw DE, Jura N, Moasser MM.
Cell Reports 2022 Feb 1;38(5):110291
Deposited structure of the HER3 pseudokinase bound to bosutinib
PDB: 6OP9
Evolution of enhanced innate immune evasion by SARS-CoV-2.
Thorne LG, Bouhaddou M, Reuschl AK, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MVX, Hosmillo M, Fossati A, Ragazzini R, Jungreis I, Ummadi M, Rojc A, Turner J, Bischof ML, Obernier K, Braberg H, Soucheray M, Richards A, Chen KH, Harjai B, Memon D, Hiatt J, Rosales R, McGovern BL, Jahun A, Fabius JM, White K, Goodfellow IG, Takeuchi Y, Bonfanti P, Shokat K, Jura N, Verba K, Noursadeghi M, Beltrao P, Kellis M, Swaney DL, García-Sastre A, Jolly C, Towers GJ, Krogan NJ.
Nature 2022 Feb;602(7897):487-495
Structures of the active HER2/HER3/NRG1b receptor complex reveal a dynamic dimer interface.
Diwanji D, Trenker R, Thaker TM, Wang F, Agard DA, Verba KA, Jura N.
Nature 2021 Dec; 600(7888):339-343
Deposited Structure of the HER2/HER3/NRG1b heterodimer
PDB: 7MN5Deposited Structure of the HER2 S310F/HER3/NRG1b heterodimer
PDB: 7MN6Deposited Structure of the HER2/HER3/NRG1b heterodimer bound to Trastuzumab
PDB: 7MN8
Highlighted in Nature Chemical Biology:
Avoiding arm contact.
Miura, G. Nat Chem Biol 18, 1 (2022)Highlighted in Cancer Discovery:
Structure of the Ligand-Bound HER2–HER3 Dimer Reveals a Dynamic Interface
Cancer Discovery 12(1): 14 (2022)
A protein network map of head and neck cancer reveals PIK3CA mutant drug sensitivity.
Swaney DL, Ramms DJ, Wang Z, Park J, Goto Y, Soucheray M, Bhola N, Kim K, Zheng F, Zeng Y, McGregor M, Herrington KA, O’Keefe R, Jin N, VanLandingham NK, Foussard H, Von Dollen J, Bouhaddou M, Jimenez-Morales D, Obernier K, Kreisberg JF, Kim M, Johnson DE, Jura N, Grandis JR, Gutkind JS, Ideker T, Krogan NJ.
Science 2021 Oct;374(6563):eabf2911
Therapeutic implications of activating noncanonical PIK3CA mutations in head and neck squamous cell carcinoma.
Jin N, Keam B, Cho J, Lee MJ, Kim HR, Torosyan H, Jura N, Ng PK, Mills GB, Li H, Zeng Y, Barbash Z, Tarcic G, Kang H, Bauman JE, Kim MO, VanLandingham NK, Swaney DL, Krogan NJ, Johnson DE, Grandis JR.
Journal of Clinical Investigation 2021 Nov 15;131(22)
Mutant HER2 needs mutant HER3 to be an effective oncogene.
Trenker R, Diwanji D, Jura N.
Cell Reports Medicine 2021 Aug 6; 2(8):100361
Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Agnew C, Ayaz P, Kashima R, Loving HS, Ghatpande P, Kung JE, Underbakke ES, Shan Y, Shaw DE, Hata A, Jura N.
Nature Communications 2021 Aug 16; 12(1):4950
Deposited structure of the BMPR2 D485G kinase in complex with ADP
PDB: 6UNPDeposited structure of the ALK2 K493A kinase in complex with AMP-PNP
PDB: 6UNQDeposited structure of the ALK2 K492A/K493A kinase in complex with AMP-PNP
PDB: 6UNRDeposited structure of the ALK2 K492A/K493A kinase in complex with LDN-193189
PDB: 6UNS
State of the structure address on MET receptor activation by HGF.
Linossi EM, Estevam GO, Oshima M, Fraser JS, Collisson EA, Jura N.
Biochem Soc Trans. 2021 Apr 30; 49(2):645-661.
A survey of the kinome pharmacopeia reveals multiple scaffolds and targets for the development of novel anthelmintics.
Knox J, Joly N, Linossi EM, Carmona-Negrón JA, Jura N, Pintard L, Zuercher W, Roy PJ.
Science Reports 2021 Apr 28; 11(1):9161.
Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Schuller M, Correy GJ, Gahbauer S, Fearon D, Wu T, Díaz RE, Young ID, Martins LC, Smith DH, Schulze-Gahmen U, Owens TW, Deshpande I, Merz GE, Thwin AC, Biel JT, Peters JK, Moritz M, Herrera N, Kratochvil HT, QCRG Structural Biology Consortium, Aimon A, Bennett JM, Neto JB, Cohen AE, Dias A, Douangamath A, Dunnett L, Fedorov O, Ferla MP, Fuchs MR, Gorrie-Stone TJ, Holton JM, Johnson MG, Krojer T, Meigs G, Powell AJ, Rack JGM, Rangel VL, Russi S, Skyner RE, Smith CA, Soares AS, Wierman JL, Zhu K, O’Brien P, Jura N, Ashworth A, Irwin JJ, Thompson MC, Gestwicki JE, von Delft F, Shoichet BK, Fraser JS, Ahel I.
Science Advances 2021 04; 7(16).
Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2.
Kushal Suryamohan, Devan Diwanji, Eric W. Stawiski, Ravi Gupta, Shane Miersch, Jiang Liu, Chao Chen, Ying-Ping Jiang, Frederic A. Fellouse, J. Fah Sathirapongsasuti, Patrick K. Albers, Tanneeru Deepak, Reza Saberianfar, Aakrosh Ratan, Gavin Washburn, Monika Mis, Devi Santhosh, Sneha Somasekar, G. H. Hiranjith, Derek Vargas, Sangeetha Mohan, Sameer Phalke, Boney Kuriakose, Aju Antony, Mart Ustav Jr, Stephan C. Schuster, Sachdev Sidhu, Jagath R. Junutula, Natalia Jura, and Somasekar Seshagiri.
Commun Biol. 2021 04 12; 4(1):475.
Drugging the ‘undruggable’ MYCN oncogenic transcription factor: Overcoming previous obstacles to impact childhood cancers.
Wolpaw AJ, Bayliss R, Büchel G, Dang CV, Eilers M, Gustafson WC, Hansen GM, Jura N, Knapp S, Lemmon MA, Levens D, Maris JM, Marmorstein R, Metallo SJ, Park JR, Penn LZ, Rape M, Roussel MF, Shokat KM, Tansey WP, Verba KA, Vos SM, Weiss WA, Wolf E, Mossé YP.
Cancer Research 2021 Jan 28.
Expression and purification of active human kinases using Pichia pastoris as a general-purpose host.
Abdel Aziz MH, Fan Y, Liu L, Moasser MM, Fu H, Jura N, Arkin MR.
Protein Expr Purif. 2021 Mar; 179:105780.
Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms.
Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, Jureka AS, Obernier K, Guo JZ, Batra J, Kaake RM, Weckstein AR, Owens TW, Gupta M, Pourmal S, Titus EW, Cakir M, Soucheray M, McGregor M, Cakir Z, Jang G, O’Meara MJ, Tummino TA, Zhang Z, Foussard H, Rojc A, Zhou Y, Kuchenov D, Hüttenhain R, Xu J, Eckhardt M, Swaney DL, Fabius JM, Ummadi M, Tutuncuoglu B, Rathore U, Modak M, Haas P, Haas KM, Naing ZZC, Pulido EH, Shi Y, Barrio-Hernandez I, Memon D, Petsalaki E, Dunham A, Marrero MC, Burke D, Koh C, Vallet T, Silvas JA, Azumaya CM, Billesbølle C, Brilot AF, Campbell MG, Diallo A, Dickinson MS, Diwanji D, Herrera N, Hoppe N, Kratochvil HT, Liu Y, Merz GE, Moritz M, Nguyen HC, Nowotny C, Puchades C, Rizo AN, Schulze-Gahmen U, Smith AM, Sun M, Young ID, Zhao J, Asarnow D, Biel J, Bowen A, Braxton JR, Chen J, Chio CM, Chio US, Deshpande I, Doan L, Faust B, Flores S, Jin M, Kim K, Lam VL, Li F, Li J, Li YL, Li Y, Liu X, Lo M, Lopez KE, Melo AA, Moss FR, Nguyen P, Paulino J, Pawar KI, Peters JK, Pospiech TH, Safari M, Sangwan S, Schaefer K, Thomas PV, Thwin AC, Trenker R, Tse E, Tsui TKM, Wang F, Whitis N, Yu Z, Zhang K, Zhang Y, Zhou F, Saltzberg D, , Hodder AJ, Shun-Shion AS, Williams DM, White KM, Rosales R, Kehrer T, Miorin L, Moreno E, Patel AH, Rihn S, Khalid MM, Vallejo-Gracia A, Fozouni P, Simoneau CR, Roth TL, Wu D, Karim MA, Ghoussaini M, Dunham I, Berardi F, Weigang S, Chazal M, Park J, Logue J, McGrath M, Weston S, Haupt R, Hastie CJ, Elliott M, Brown F, Burness KA, Reid E, Dorward M, Johnson C, Wilkinson SG, Geyer A, Giesel DM, Baillie C, Raggett S, Leech H, Toth R, Goodman N, Keough KC, Lind AL, , Klesh RJ, Hemphill KR, Carlson-Stevermer J, Oki J, Holden K, Maures T, Pollard KS, Sali A, Agard DA, Cheng Y, Fraser JS, Frost A, Jura N, Kortemme T, Manglik A, Southworth DR, Stroud RM, Alessi DR, Davies P, Frieman MB, Ideker T, Abate C, Jouvenet N, Kochs G, Shoichet B, Ott M, Palmarini M, Shokat KM, García-Sastre A, Rassen JA, Grosse R, Rosenberg OS, Verba KA, Basler CF, Vignuzzi M, Peden AA, Beltrao P, Krogan NJ.
Science. 2020 12 04; 370(6521).
The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Titus EW, Deiter FH, Shi C, Wojciak J, Scheinman M, Jura N, Deo RC.
Nat Struct Mol Biol. 2020 12; 27(12):1142-1151.
An International Multicenter Evaluation of Inheritance Patterns, Arrhythmic Risks, and Underlying Mechanisms of CASQ2-Catecholaminergic Polymorphic Ventricular Tachycardia.
Ng K, Titus EW, Lieve KV, Roston TM, Mazzanti A, Deiter FH, Denjoy I, Ingles J, Till J, Robyns T, Connors SP, Steinberg C, Abrams DJ, Pang B, Scheinman MM, Bos JM, Duffett SA, van der Werf C, Maltret A, Green MS, Rutberg J, Balaji S, Cadrin-Tourigny J, Orland KM, Knight LM, Brateng C, Wu J, Tang AS, Skanes AC, Manlucu J, Healey JS, January CT, Krahn AD, Collins KK, Maginot KR, Fischbach P, Etheridge SP, Eckhardt LL, Hamilton RM, Ackerman MJ, Noguer FRI, Semsarian C, Jura N, Leenhardt A, Gollob MH, Priori SG, Sanatani S, Wilde AAM, Deo RC, Roberts JD.
Circulation 2020 Sep 08; 142(10):932-947.
A SARS-CoV-2 protein interaction map reveals targets for drug repurposing.
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ.
Nature 2020 07; 583(7816):459-468.
Receptor tyrosine kinase activation: From the ligand perspective.
Trenker R, Jura N.
Curr Opin Cell Biol. 2020 Apr;63:174-185.
The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.
Agnew C, Liu L, Liu S, Xu W, You L, Yeung W, Kannan N, Jablons D, Jura N.
Journal of Biological Chemistry 2019 Sep 13;294(37):13545-13559.
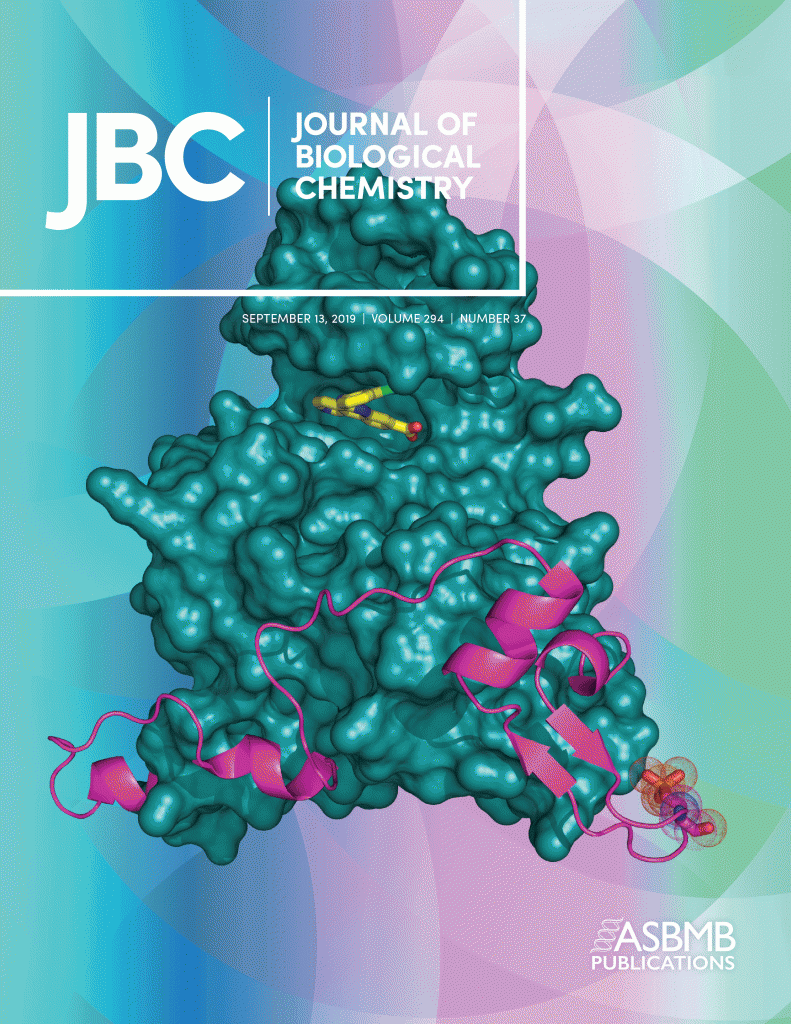
Editor’s Pick in Science Signaling:
New connections: HIPK2 becomes crystal clear.
Foley JF
Science Signaling 30 Jul 2019: Vol12, Issue 592, eaay8936
Editor’s highlight in JBC:
The long-awaited structure of HIPK2.
Murphy JM
J Biol Chem. 2019 Sep 13;294(37):13560-13561.
Deposited structure of the HIPK2 kinase in complex with an inhibitor CX-4945:
PEAK3/C19orf35 pseudokinase, a new NFK3 kinase family member, inhibits CrkII through dimerization.
Lopez ML, Lo M, Kung JE, Dudkiewicz M, Jang GM, Von Dollen J, Johnson JR, Krogan NJ, Pawłowski K, Jura N.
Proc Natl Acad Sci U S A 2019 Jul 30;116(31):15495-15504.
More than the sum of the parts: Toward full-length receptor tyrosine kinase structures.
Diwanji D, Thaker T, Jura N.
IUBMB Life. 2019 Jun;71(6):706-720. Review.
Prospects for pharmacological targeting of pseudokinases.
Kung JE, Jura N.
Nature Reviews Drug Discovery 2019 Jul;18(7):501-526. doi: 10.1038/s41573-019-0018-3. Review.
Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Ruiz K, Thaker TM, Agnew C, Miller-Vedam L, Trenker R, Herrera C, Ingaramo M, Toso D, Frost A, Jura N.
Nature Communications 2019 Jan 31;10(1):531.
Deposited structure of PGAM5 dodecamer:
Deposited structure of PGAM5 H105A dimer:
The pseudokinase TRIB1 toggles an intramolecular switch to regulate COP1 nuclear export.
Kung JE, Jura N.
EMBO Journal 2019 Feb 15;38(4).
Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations.
Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S.
Cancer Cell 2018 Nov 12;34(5):792-806.
Activating HER3 mutations in breast cancer.
Mishra R, Alanazi S, Yuan L, Solomon T, Thaker TM, Jura N, Garrett JT.
Oncotarget 2018 Jun 12;9(45):27773-27788.
Phosphorylated EGFR Dimers Are Not Sufficient to Activate Ras.
Liang SI, van Lengerich B, Eichel K, Cha M, Patterson DM, Yoon TY, von Zastrow M, Jura N, Gartner ZJ.
Cell Reports 2018 Mar 6;22(10):2593-2600. doi: 10.1016/j.celrep.2018.02.031.
Regulation of Kinase Activity in the Caenorhabditis elegans EGF Receptor, LET-23.
Liu L, Thaker TM, Freed DM, Frazier N, Malhotra K, Lemmon MA, Jura N.
Structure 2018 Feb 6;26(2):270-281.e4.
Deposited structure of the LET-23 kinase domain
Feedback regulation of RTK signaling in development.
Neben CL, Lo M, Jura N, Klein OD.
Developmental Biology 2019 Mar 1;447(1):71-89. Review.
Switching on BTK-One Domain at a Time.
Agnew C, Jura N.
Structure 2017 Oct 3;25(10):1469-1470.
EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms.
van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N.
Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):E2836-E2845.
Structural Basis for the Non-catalytic Functions of Protein Kinases.
Kung JE, Jura N.
Structure 2016 Jan 5;24(1):7-24. Review.
Src defines a new pool of EGFR substrates.
Michael N, Jura N.
Nature Structural and Molecular Biology 2015 Dec;22(12):945-7.
Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Kovacs E, Das R, Wang Q, Collier TS, Cantor A, Huang Y, Wong K, Mirza A, Barros T, Grob P, Jura N, Bose R, Kuriyan J.
Molecular and Cellular Biology 2015 Sep 1;35(17):3083-102.
Deposited structure of the EGFR I682Q mutant
Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Littlefield P, Liu L, Mysore V, Shan Y, Shaw DE, Jura N.
Science Signaling 2014 Dec 2;7(354):ra114.
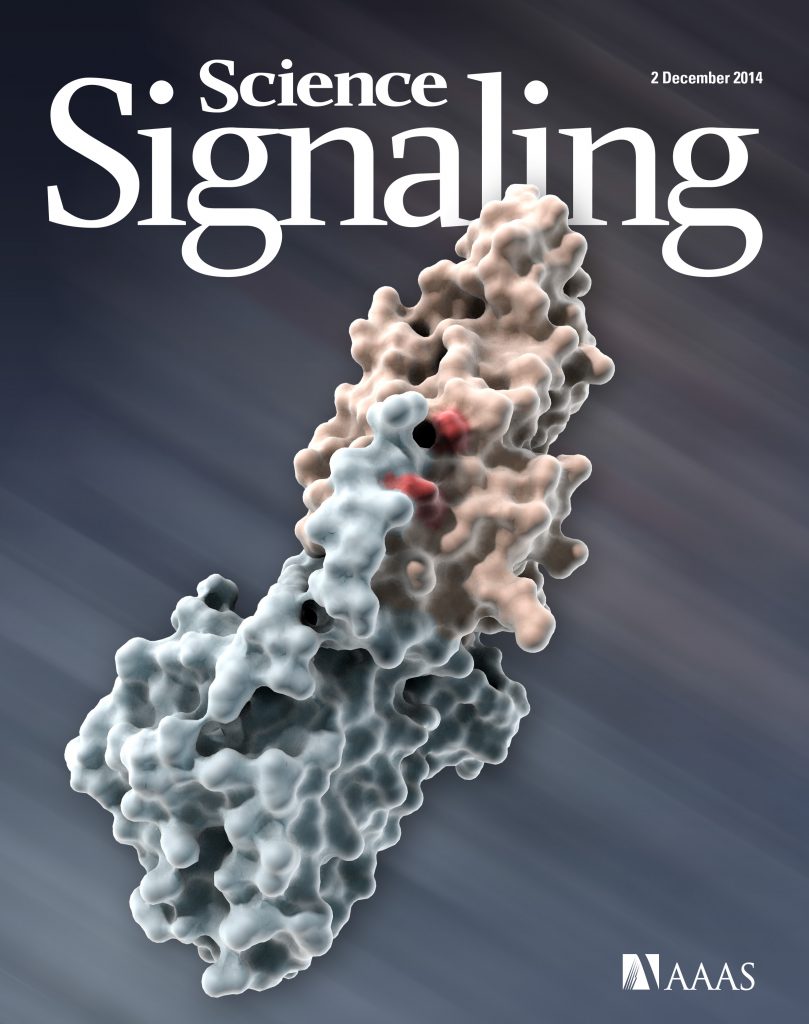
Deposited structure of the EGFR/HER3 kinase heterodimer:
Deposited structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-Q790R mutation:
Deposited structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-E909G mutation:
EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma.
Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA, Chen J, Lau J, Knobbe-Thomsen C, Weller M, Jura N, Reifenberger G, Shokat KM, Weiss WA.
Cancer Cell 2013 Oct 14;24(4):438-49.
A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties.
Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, Lucet IS.
Biochemical Journal 2014 Jan 15;457(2):323-34.
EGFR lung cancer mutants get specialized.
Littlefield P, Jura N.
Proc Natl Acad Sci U S A. 2013 Sep 17;110(38):15169-70.
Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms.
Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J.
Molecular Cell 2011 Apr 8;42(1):9-22. Review.
Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J.
Proc Natl Acad Sci U S A. 2009 Dec 22;106(51):21608-13.
Deposited structure of the HER3 kinase domain in complex with AMP-PNP:
Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J.
Cell 2009 Jun 26;137(7):1293-307.
Preview in Cell:
The juxtamembrane region of EGFR takes center stage.
Hubbard SR.
Cell 2009 Jun 26;137(7):1181-3.
Deposited structure of the inactive EGFR V924R kinase dimer:
Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J.
Nature 2007 Nov 29;450(7170):741-4.
Deposited structures of the EGFR/Mig6 complexes:
Mapping cellular routes of Ras: a ubiquitin trail.
Jura N, Bar-Sagi D.
Cell Cycle 2006 Dec;5(23):2744-7. Review.
A mouse model of hereditary pancreatitis generated by transgenic expression of R122H trypsinogen.
Archer H, Jura N, Keller J, Jacobson M, Bar-Sagi D.
Gastroenterology 2006 Dec;131(6):1844-55.

Editorial in Gastroenterology:
Genetically defined models of chronic pancreatitis.
Schmid RM, Whitcomb DC.
Gastroenterology 2006 Dec;131(6):2012-5.
Differential modification of Ras proteins by ubiquitination.
Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D.
Molecular Cell 2006 Mar 3;21(5):679-87.
Editorial in Cancer Cell:
Ras ubiquitination: coupling spatial sorting and signal transmission.
Rodriguez-Viciana P, McCormick F.
Cancer Cell 2006 Apr;9(4):243-4.
Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis.
Mezyk-Kopec R, Bzowska M, Potempa J, Bzowska M, Jura N, Sroka A, Black RA, Bereta J.
Infection and Immunity 2005 Mar;73(3):1506-14.
An ex vivo model for functional studies of myofibroblasts.
Kilarski WW, Jura N, Gerwins P.
Laboratory Investigation 2005 May;85(5):643-54.
Chronic pancreatitis, pancreatic adenocarcinoma and the black box in-between.
Jura N, Archer H, Bar-Sagi D.
Cell Research 2005 Jan;15(1):72-7. Review.
Tumour necrosis factor-alpha stimulates expression of TNF-alpha converting enzyme in endothelial cells.
Bzowska M, Jura N, Lassak A, Black RA, Bereta J.
European Journal of Biochemistry 2004 Jul;271(13):2808-20.
Inactivation of Src family kinases inhibits angiogenesis in vivo: implications for a mechanism involving organization of the actin cytoskeleton.
Kilarski WW, Jura N, Gerwins P.
Experimental Cell Research 2003 Nov 15;291(1):70-82.
hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl.
Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D.
Current Biology 2003 Feb 18;13(4):308-14.
In the news:
Split personalities: the agonistic antagonist Sprouty.
Christofori G.
Nature Cell Biology 2003 May;5(5):377-9.LeBrasseur N.
Nature Cell Biology 2003 May;5(5):377-9.